Dear Sergey !
As above mentioned I implemented test code and evaluated random 9 of 300 know pair of validated measurements:
The result improved greatly (individual concentrations never goes below zero) - but not perfect yet (sum of concentration near to 1):
#Eval ∑Concentrations (must be 1)
1 0,954
7 1,017
33 1,033
51 1,021
100 1,003
153 1,044
200 1,041
250 0,924
298 0,995
If I check the details in individual concentration components level: put exactly the same input as model built (see above mentioned Eval #1, 7, .. #298) to mlpprocess and compare the two outputs: original at train set and output of mlpprocess I have some differences, just one examples -see attached picture file.
Attachment:
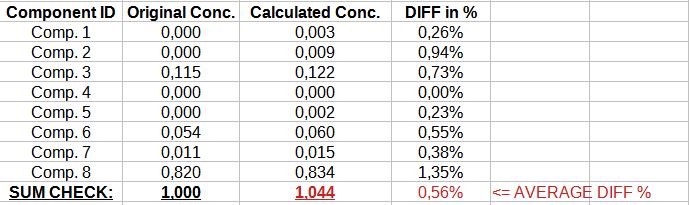 ALGLIB Sergey mlpprocess one example Concentrations..JPG [ 46.71 KiB | Viewed 34939 times ]
ALGLIB Sergey mlpprocess one example Concentrations..JPG [ 46.71 KiB | Viewed 34939 times ]
What do you think : is it the expected level of quality what I can get or is there any tips and tricks / room to improve the quality of the result?
Thanks in advance,
Best Regards,
Péter



